Input and output description
Input files
FASTA files of coding regions (CDS) or of coding transcripts for each species in the dataset.
GFF3 file for the focal species, allows collinearity (syntenic) analysis and anchor pair paralog KS distribution.
ksrates configuration file(s) (for more details see Configuration files).
Output files and directory organization
Note
In the following listings of directory and file names, species is used as a placeholder for the actual (informal) name of the focal species (e.g. elaeis) as specified in the ksrates configuration file.
Main output
rate_adjustment/species: this directory collects the output files of the substitution rate-adjustment relative to the focal species.Figures:
Rate-adjusted mixed paralog–ortholog KS distribution plot in PDF format (
mixed_species_adjusted.pdf).Input phylogenetic tree in PDF format with branch length set to KS distances estimated from ortholog KS distributions (
tree_species_distances.pdf).Rate-adjusted mixed anchor pair–ortholog KS distribution clustered for inference of putative WGDs, with only significant clusters retained (
mixed_species_anchor_clusters.pdf).Rate-adjusted mixed paralog–ortholog KS distribution with superimposed exponential-lognormal mixture model inference of putative WGDs (
mixed_species_elmm.pdf).Rate-adjusted mixed paralog– and anchor pair–ortholog KS distributions with superimposed lognormal-only mixture model for inference of putative WGDs (
mixed_species_lmm_paranome.pdfandmixed_species_lmm_colinearity.pdf).Multi-panel figure(s) of the ortholog KS distributions used to adjust a divergent species pair (
orthologs_species1_species2.pdf).Unadjusted naive mixed paralog–ortholog KS distribution plot in PDF format (
mixed_species_unadjusted.pdf).Original input phylogenetic tree in PDF format with fixed branch lengths (
tree_species.pdf).
Files:
Raw rate-adjustment results for each trio (
adjustment_table_species_all.tsv). Tabular format.Each row shows the result for a species pair (column 2
Focal_Speciesand 3Sister_Species) diverging at a certain node (column 1Node) and adjusted with the outgroup in column 3Out_Species. The rate-adjusted mode with associated standard deviation are given in column 4Adjusted_Modeand 5Adjusted_Mode_SD; for comparison the unadjusted original mode with associated standard deviation is provided in column 6Original_Modeand 7Original_Mode_SD. The branch-specific KS contributions for the divergent species pair are listed in column 8Ks_Focaland 9Ks_Sister; the KS`distance of the outgroup to the divergence event of the species pair is listed in column 10 ``Ks_Out`.
Raw rate-adjustment results on a divergent pair using four outgroups.
Final rate-adjustment results for each divergent species pair after finding a consensus value in case of multiple outgroups (
adjustment_table_species.tsv). Tabular format.Each row shows the result for a species pair (column 2
Focal_Speciesand 3Sister_Species) diverging at a certain node (column 1Node). Columns 4–7 report the consensus obtained by taking the mean of multiple outgroups (if available): rate-adjusted mode with standard deviation in column 4Adjusted_Mode_Meanand 5Adjusted_Mode_SD_Mean, branch-specific KS contributions for the divergent species pair in column 6Ks_Focal_Meanand 7Ks_Sister_Mean. Columns 8–11 report the consensus obtained when considering only the best outgroup: rate-adjusted mode with standard deviation in column 8Adjusted_Mode_Bestand 9Adjusted_Mode_SD_Best, KS contributions for the divergent species pair in column 10Ks_Focal_Bestand 11Ks_Sister_Best. For comparison the unadjusted original mode with associated standard deviation is provided in column 12Original_Modeand 13Original_Mode_SD.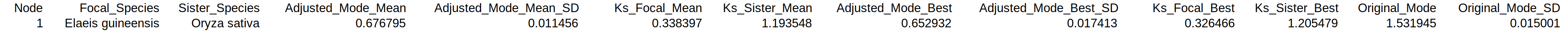
Consensus result for the divergent pair obtained from the four raw rate-adjustments.
Original input phylogenetic tree in ASCII format and list of sister species and outgroup species per node (
tree_species.txt).List of trios used for substitution rate-adjustment (
ortholog_trios_species.tsv).List of species pairs for which ortholog KS distributions are estimated using wgd (
ortholog_pairs_species.txt).
rate_adjustment/species/paralogs_analyses: this directory collects secondary output files produced during the inference of putative WGD signals through mixture modeling (see Mixture modeling of paralog KS distributions).From anchor KS clustering:
Anchor pair KS distribution with highlighted clusters of segment pair medians (
anchor_clusters_species_medians.pdf).Rate-adjusted mixed anchor pair–ortholog KS distributions clustered for inference of putative WGDs, with all inferred clusters (
mixed_species_anchor_clusters_unfiltered.pdf).
From exponential-lognormal mixture modeling:
Plots showing the kernel density estimation (KDE) and spline obtained from the log-transformed whole-paranome KS distribution (
elmm_species_kde_spline.pdf).Plots showing the peaks detected in the spline (
elmm_species_peaks.pdf).Multi-panel figure showing fitted mixture models obtained with data-driven and hybrid initializations (
elmm_species_models_data_driven.pdf).Multi-panel figure showing the best-fitted mixture model obtained for each number of components with random initialization (
elmm_species_models_random.pdf).TSV and TXT files collecting component parameters (
elmm_species_parameters.tsvandelmm_species_parameters.txt) (see Exponential-lognormal mixture model for more details on the file format).
From lognormal-only mixture modeling:
Multi-panel figure showing the best-fitted mixture model on whole-paranome and anchor pair KS distributions obtained for each number of components (
lmm_species_all_models_paranome.pdfandlmm_species_all_models_colinearity.pdf).TSV and TXT files collecting component parameters (
lmm_species_parameters_colinearity.tsv,lmm_species_parameters_colinearity.txt,lmm_species_parameters_paranome.tsvandlmm_species_parameters_paranome.txt) (see Lognormal mixture model for more details on the file format).
Nextflow log files
rate_adjustment/species/log_XXXXXXXX: when launching ksrates as a Nextflow pipeline, each execution generates a log directory named with a unique 8-character ID stated at the beginning of a Nextflow run. Details about how the processes of the workflow are proceeding and about encountered warnings or errors are stored in log files collected in this directory:setup_adjustment.logshows the progress in checking input files and setting up species trios and pairs for rate-adjustment.wgd_paralogs.logshows the progress in estimating paralog KS values.set_orthologs.logstates whether ortholog KS data are already available or are missing for each species pair.estimate_peak.logshows the progress in updating the ortholog KS databases from already existing ortholog KS data.wgd_orthologs_species1_species2.logshows the progress in estimating ortholog KS values for a species pair.plot_ortholog_distributions.logshows the progress in plotting the ortholog KS distributions.rate_adjustment.logshows the progress in performing the actual rate-adjustment step.paralogs_analyses.logshows the progress in analyzing the paralog distribution to detect potential WGD signatures through anchor KS clustering, exponential-lognormal mixture modeling and/or lognormal-only mixture modeling.
KS estimate output (wgd)
paralog_distributions/wgd_species: this directory contains the files generated during the wgd paralog KS estimation run for the focal species:species.blast.tsvlists the paralog BLAST homology hits in tabular output format (-outfmt 6)species.mcl.tsvlists the paralog gene families, one family per line from the largest to the smallest family with the gene IDs of individual family members separated by tabs.species.ks.tsvandspecies.ks_anchors.tsvare tabular format files listing the KS estimate (column 9Ks) for every paralog and anchor pair found, respectively. Other noteworthy data per pair includes the alignment coverage, identity and length (columns 2–5:AlignmentCoverage,AlignmentIdentity,AlignmentLengthandAlignmentLengthStripped), the gene family (column 7Family), the node in the gene family’s tree (column 10Node), and the weight associated with the pair’s KS estimate (column 15WeightOutliersExcluded). For more details, see the wgd documentation.
File section showing the structure of the
.ks.tsvformat.species_i-adhore: this directory contains the i-ADHoRe output files necessary for the anchor KS clustering (see Anchor KS clustering).
ortholog_distributions/wgd_species1_species2: these directories contain the files generated during the wgd one-to-one ortholog KS estimation for each species pair:species1_species2.blast.tsvlists the ortholog BLAST homology hits.Note
When the wgd ortholog KS estimation analysis is finished it is possible to delete this file to save disk space.
species1_species2.orthologs.tsvlists the one-to-one ortholog (i.e. the reciprocal best BLAST hits) between the two species, one ortholog pair per line.species1_species2.ks.tsvlists the KS estimate (column 9Ks) for every one-to-one ortholog pair found. The tabular file format is identical to the paralog.ks.tsvfile described above. However, the gene family, tree node and weight columns can be ignored since each ortholog “family” is composed of only two members.
Other output
Generated directly in the directory from where ksrates is launched:
ortholog_peak_db.tsvis a tabular data file storing the KS mode estimate (column 4Mode) and associated standard deviation (column 5Mode_SD) of the ortholog KS distribution of species pairs (columns 1–3). File name and location can be customised in the ksrates configuration file.
Structure of ortholog KS mode database.
ortholog_ks_list_db.tsvis a tabular data file storing the ortholog KS value lists (column 4Ks_Values) of species pairs (columns 1–3). File name and location can be customised in the ksrates configuration file.
Structure of ortholog KS list database.
wgd_runs_species.txtcontains a list of ksrates commands to launch the wgd paralog and ortholog analysis when using the manual pipeline (see Run example case as a manual pipeline). Note that this file is not generated if using the ksrates Nextflow pipeline.work: when using the ksrates Nextflow pipeline this directory is automatically generated by Nextflow to handle process organization and communication between processes (for more details, see the Nextflow documentation, e.g. here).
Note on wgd output files
If a ksrates Nextflow pipeline run is prematurely interrupted for some reasons (e.g. cancelled by the user or crashed) while one or more wgd runs were still ongoing, the latter will leave temporary directories and incomplete files within paralog_distributions and/or ortholog_distributions (e.g. BLAST files). Such leftovers are by default automatically detected and removed at the end of the workflow as a safety measure to avoid that the next run continues the task from incomplete data.
It is possible to preserve the leftover files for investigating what caused the pipeline to crash (see preserve parameter in Nextflow configuration file). In this case it will be later necessary to manually remove the leftovers before relaunching the pipeline, otherwise the workflow will immediately stop and return an error message in the Nextflow log files (wgd_paralogs.log and/or wgd_orthologs_species1_species2.log).