FAQs about ksrates
Nextflow
If Nextflow crashes, where do I look for the error message?
When Nextflow crashes, an error box appears in the terminal and shows which process stopped the pipeline, the error message lines and points to complete output files. For example:
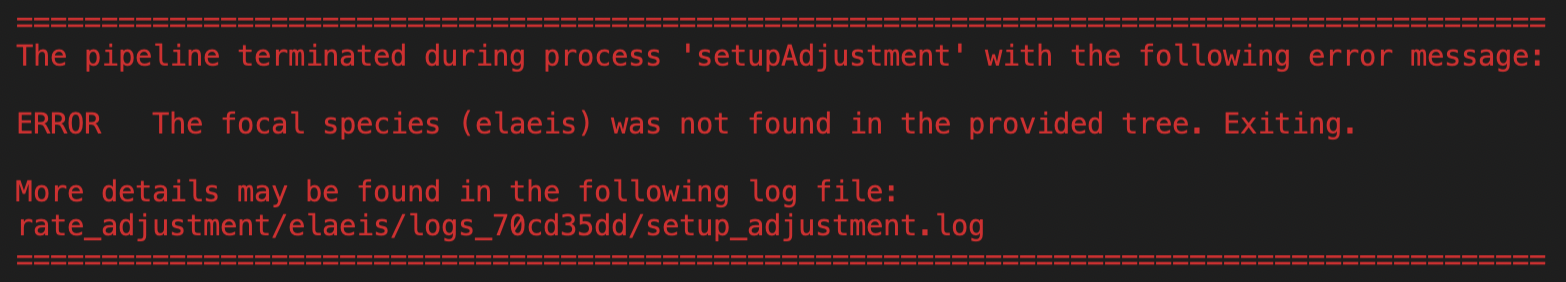
Here the error is traced back to the setupAdjustment Nextflow process and it is due to a wrong setting in the ksrates configuration file concerning the input phylogeny.
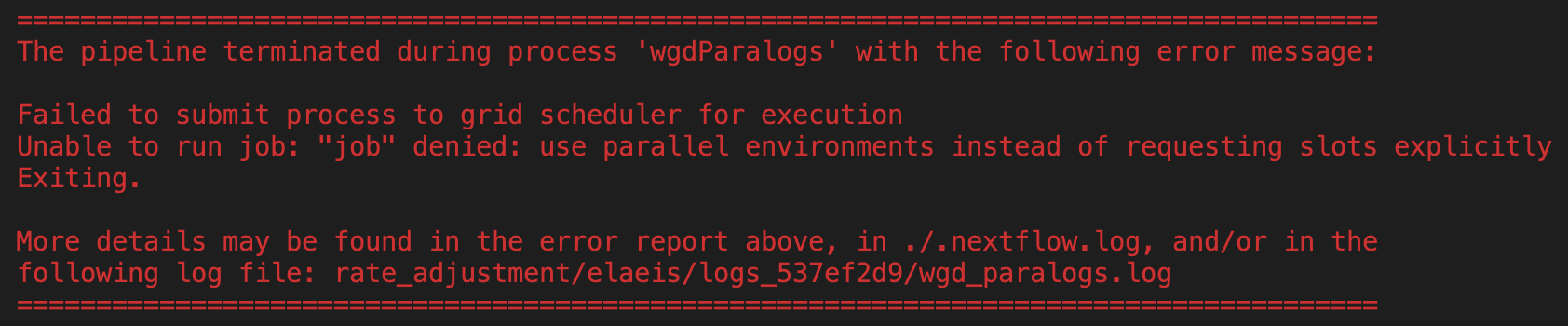
Here the error is traced back to the wgdParalogs Nextflow process and it is due to a wrong setting in the Nextflow configuration file concerning communication with the computer cluster.
More details about any error that terminated the pipeline can usually be found in log files that are stored in rate_adjustment/<focal_species>/log_XXXXXXXX. The log folder name is unique for each run and is reported on the terminal in the error box and also at the beginning and end of a Nextflow run. The more general Nextflow log file .nextflow.log in the folder where the pipeline has been launched could also contain additional information.
I am getting a failed to reserve page summary memory error?
Some cluster configurations may not be fully compatible with certain built-in Nextflow process directives used in the Nextflow configuration file. For example, on some cluster configurations the ksrates pipeline fails with a failed to reserve page summary memory error when using the built-in memory directive to define how much memory a process requires. In these cases it is likely that instead the clusterOptions process directive (see the Nextflow documentation) needs to be used to define configuration settings specific to your cluster. We advise to contact your cluster administrator about these.
How do I update a previously downloaded ksrates pipeline if a new version becomes available?
nextflow pull VIB-PSB/ksrates
Containers
Containers take up quite some storage space, is this normal?
Containers are known to be voluminous, especially Docker ones.
To keep Docker images, containers and volumes under control, you can delete the ones you don’t need anymore by using the following commands:
Selectively delete images (
vibpsb/ksrates):docker image rm [image_ID]Delete all containers/images and networks that are not in use:
docker system prune -a
To remove a Singularity container, remove its .img file. You can also check and clean the Singularity cache with:
singularity cache clean [--dry-run]
General errors and warnings
The KS analysis step seems stuck.
KS analysis can take up to several hours, depending on the size of the genome, the size of the gene families, available resources, etc., and can look frozen when it doesn’t output any of the ongoing operations. The building of the BLAST database and the gene family KS analyses can take quite some time. Particularly for paralog KS analyses, it can happen that even if the last gene family has been processed the analysis of more complex earlier gene families is still not complete.
What does “No codeml results for gene family [ID]” mean?
The Nextflow wgd_paralogs.log file reports details over the KS analysis from the paralog or ortholog gene families. No codeml results for gene family [ID]... means that no KS estimates could be produced for that gene family.
Configuration
How can I change the fit of the KDE line(s) on the paranome and anchor distributions?
Adjust the kde_bandwidth_modifier parameter in the expert configuration file (see Expert configuration file).